xGen™ 2S DNA Library Prep
High library complexity from damaged and ultra-low inputs
xGen 2S DNA Library Prep Kits enable you to build high complexity libraries from a wide range of input quantities and a variety of sample types. This kit was design for both PCR and PCR-Free workflows and is ideal for ChIP Seq and Hi-C.
xGen NGS—made for challenging samples.
Ordering
xGen 2S DNA Library Prep kits
- Ideal for processing damaged samples such as low cell number ChIP and Hi-C enriched samples
- Highly efficient library preparation workflow supports ultra-low inputs as low as 10 pg
- No adapter titration so maintains ligation efficiency at low input quantities
- High ligation efficiency with low artifact formation
- Same great Swift Accel-NGS 2S DNA library prep rebranded as IDT xGen
- Designed for easy automation
Product details
Legacy Swift Accel-NGS 2S DNA Library Kits are now available as the xGen 2S DNA Library Prep Kit. The xGen 2S DNA Library Prep Kit prepares high complexity NGS libraries from damaged and low input double stranded (dsDNA) samples that have undergone DNA fragmentation. The workflow is designed to support a broad range of input quantities (10 pg to 1 µg) for whole genome and metagenome sequencing, direct sequencing of enriched samples such as ChIP and Hi-C, or targeted sequencing by hybridization capture. Unique end repair capabilities of 5’ and 3’ termini improve ligation efficiency of damaged samples while sequential ligation overcomes the requirement for adapter titration, thereby maintaining efficient ligation of low nanogram and picogram input quantities.
Workflow options
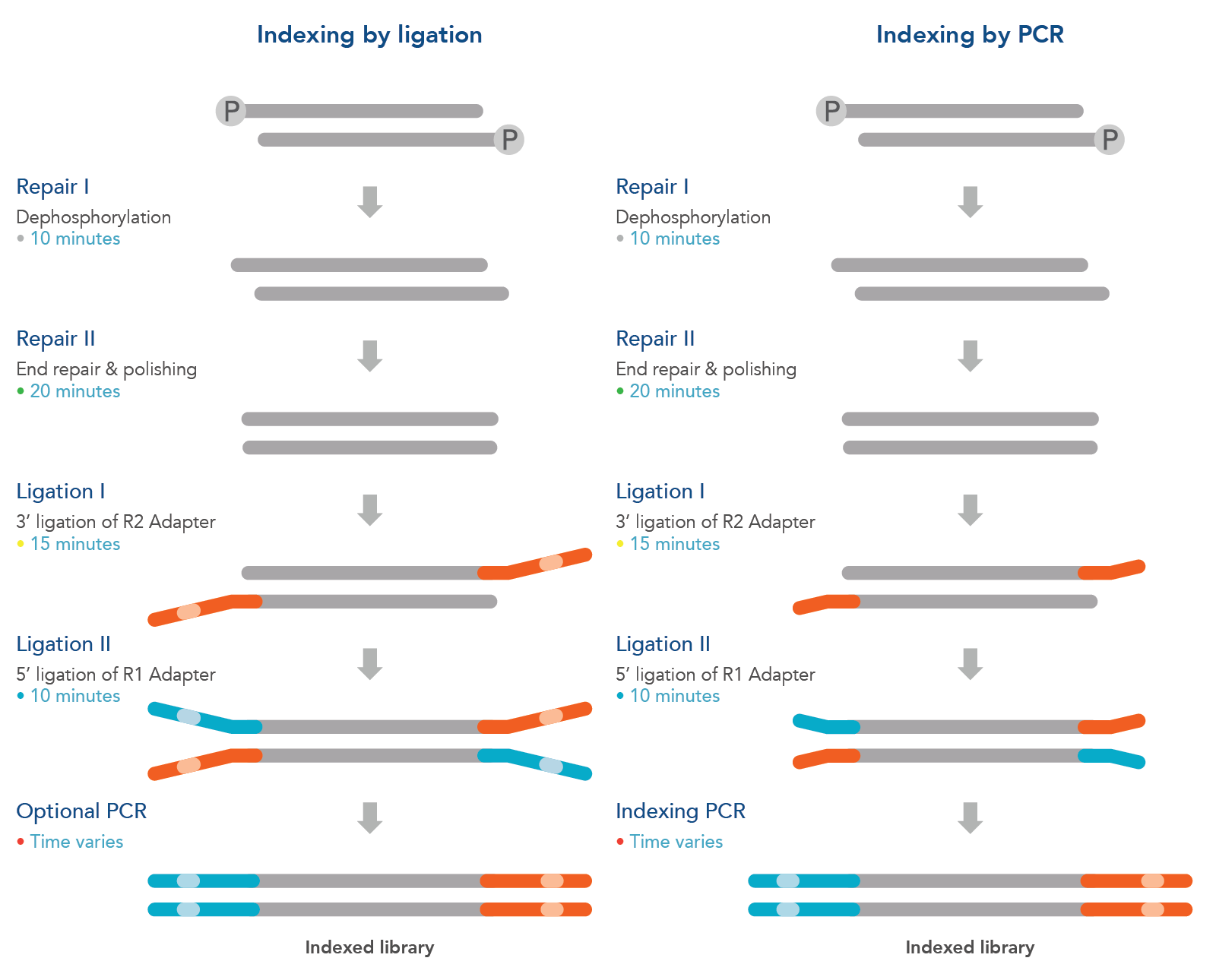
Indexing by ligation workflow:
The xGen 2S DNA Library Prep Kit has an indexing by ligation workflow compatible with xGen 2S Full-Length Adapters for Illumina® sequencing (supplied separately). This workflow supports PCR-free sequencing from a minimum DNA input of 100 ng, or if using lower inputs down to the picogram range, PCR amplification can be used to increase library yield. The indexing by ligation workflow can also be used to incorporate molecular identifier (MID) indexed adapters for subsequent error correction during data analysis. The MID adapters enable up to 96-plex multiplexed sequencing. With PCR, this workflow is compatible with the Normalase™ and Pre-Hyb Normalase Modules using the included R5 primers.
Indexing by PCR workflow:
The xGen 2S DNA Library Prep Kit also has an indexing by PCR workflow. This workflow is compatible with xGen TruY2 and TruB2 truncated adapters attached during the ligation steps. This is followed by library amplification using xGen Indexing Primers to complete the adapter sequences and add sample-specific indexes (truncated adapters and indexing primers supplied separately, see ordering information). This workflow enables up to 1536-plex multiplexed sequencing and is compatible with the Normalase and Pre-Hyb Normalase Modules using Normalase Indexing primers.
Supported applications and sample types
The xGen 2S DNA Library Prep Kit is suitable:
- Damaged DNA samples* such as DNA extracted from chemically fixed / cross-linked cells
- Enriched DNA from low cell number ChIP and Hi-C processing
- Low input environmental / metagenomic DNA samples
- Limiting DNA samples for oncology or other research studies
- Direct sequencing of DNA libraries or targeted sequencing using hybridization capture following library preparation †
- PCR-free sequencing
* For heavily nicked or denatured samples, use the xGen ssDNA and Low Input DNA Library Kit for best results
† For hybridization capture, perform library amplification with a polymerase that can support up to 1 µg library yield
Table 1. Adapter compatibility.
| Indexing by Ligation | Indexing by PCR | |
|---|---|---|
| xGen 2S MID Adapters | ✔ | |
| xGen 2S Truncated Adapters | ✔ ‡ | |
| xGen Normalase UDI Primers | ✔ | |
| xGen Normalase CDI Primers | ✔ | |
| xGen UDI Primer Pairs | ✔ |
‡ xGen 2S Truncated Adapters (Catalog Number 10009908) are required for use with indexing primers.
Molecular Identifiers for xGen 2S Library Kits
The xGen 2S Molecular Identifiers (MIDs) are designed and optimized for use with the xGen 2S DNA Library Kit. The use of MIDs improves the detection of low frequency variants by distinguishing low frequency mutations from errors generated during PCR amplification and sequencing. Additionally, these strand-specific MIDs allow accurate filtering of PCR duplicates from fragmentation and strand duplicates. Accurate identification of duplicate reads is especially useful with samples that have undergone non-random fragmentation or are sequenced as single read. Within cfDNA or ChIP libraries, uniquely derived fragments of the same sequence and length may exist in the final dataset. Previously, these unique fragments would be identified as PCR duplicates. For more information on the use of MIDs for error correction in data processing, contact us.
Features
- Incorporate molecular identifiers
- Retain more sequencing data
- Error correction for low frequency variant calling
Pico-Scale ChIP Seq
xGen™ 2S DNA Library Prep technology enables the use of ultralow input samples for ChIP-Seq applications by utilizing a unique chemistry for the most efficient preparation of NGS libraries. xGen™ 2S DNA Library Prep Kits allow you to push the lower limits of input for your ChIP-seq applications by efficiently converting low input DNA into highly diverse and complex libraries. The proprietary chemistry, which polishes both the 5’ and 3’ ends, produces libraries with an unsurpassed conversion efficiency and balanced representation of both AT-/GC-rich regions. It is possible to construct libraries from as little as 1,000 cells, minimizing the impact of cellular heterogeneity on genome-wide epigenetic profiles for chromatin proteins and transcription factors. By constructing a more complex NGS library, xGen™ 2S DNA Library Prep Kits enables the use of low input samples for ChIP-Seq applications.
Hybridization capture
The xGen 2S DNA Library Prep Kit were developed for construction of high complexity libraries from low inputs for hybridization and bait capture applications. The kit is ideal for exome sequencing with DNA isolated from FFPE samples. The 5’ and 3’ steps repair the damaged DNA from the FFPE samples, enabling the construction of high-quality libraries from nicked, degraded, and damaged samples. xGen™ 2S DNA Library Prep Kits works with xGen™ Exome Hyb Panel and smaller panels. Indexing kits are available that allow for multiplexing samples within a lane on Illumina sequencers such as the HiSeq.
Resources
Frequently asked questions
What is the shelf life and recommended storage conditions for xGen™ 2S DNA Library Prep Kit?
The shelf life of the xGen 2S DNA Library Prep Kit is 3 months from the time the product is delivered when stored at −20ºC and handled according to the protocol. The enzymes provided in these kits are temperature sensitive so appropriate care should be taken during handling and storage.
Upon receipt, immediately store the kit at −20°C.
Can the xGen™ 2S DNA Library Prep Kit be used upstream of hybridization capture workflows?
What is the DNA input requirements for the xGen™ 2S DNA Library prep Kit?
What is the new name of the Swift Accel-NGS 2S library prep kit?
The xGen™ 2S DNA Library Prep Kit was previously known as the Swift Accel-NGS 2S DNA Library Kits before IDT acquired Swift Biosciences.